spark is a framework for big data calculation
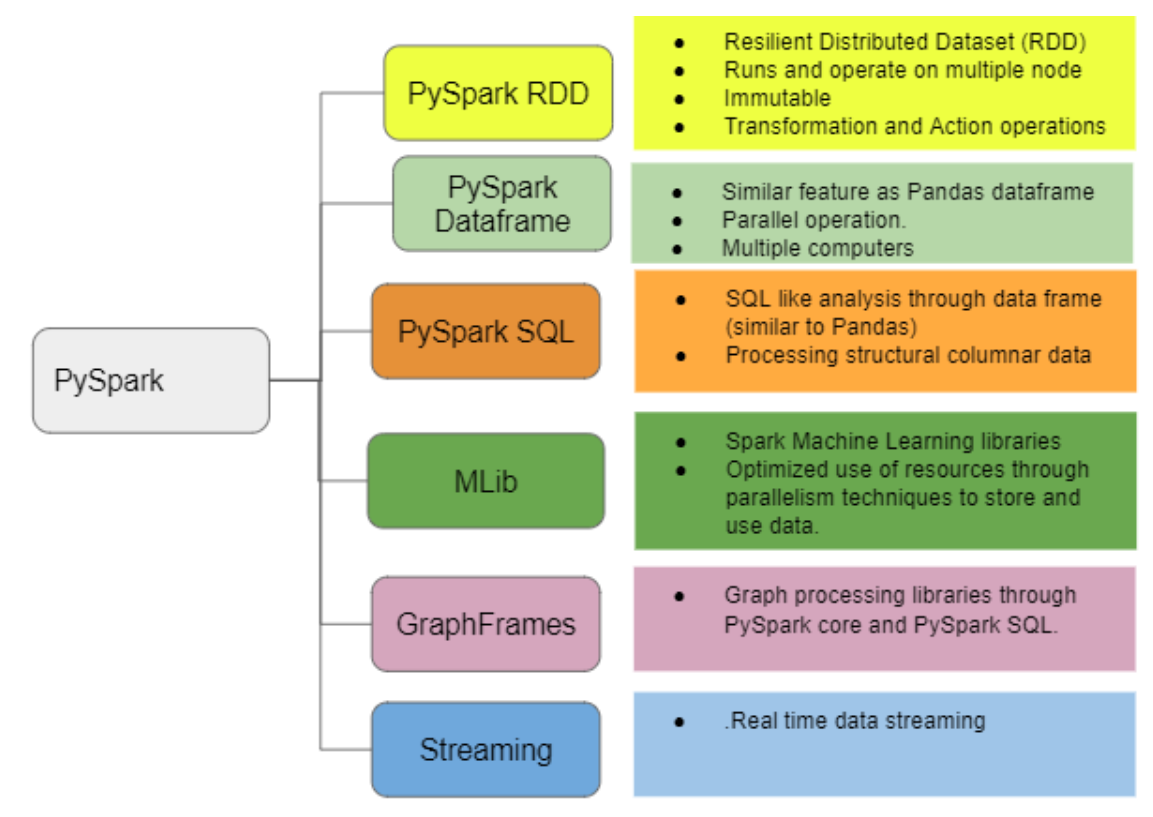 the biggest value addition in Pyspark is the parallel processing of a huge dataset on more than one computer
the biggest value addition in Pyspark is the parallel processing of a huge dataset on more than one computer
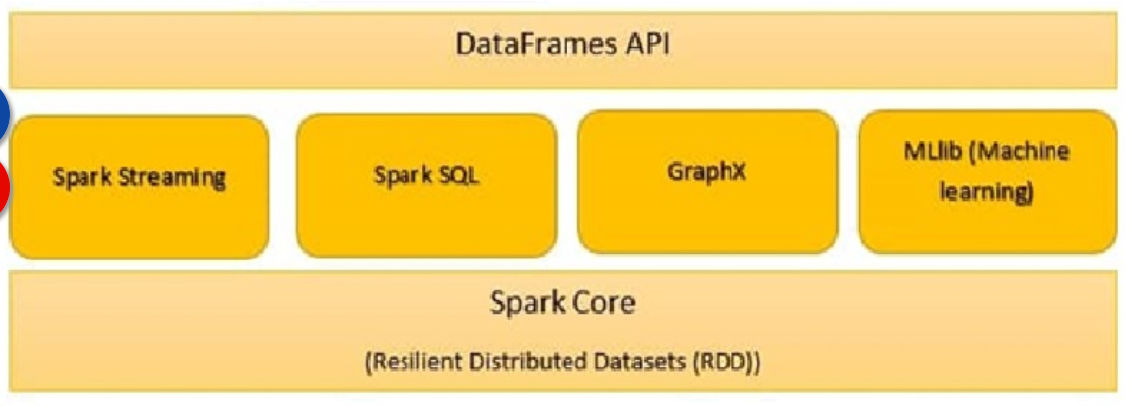 All the functionalities being provided by Apache Spark are built on the top of Spark Core. It manages all essential I/O functionalities. It is used for task dispatching and fault recovery. Spark Core is embedded with a special collection called RDD (Resilient Distributed Dataset). RDD is among the abstractions of Spark. Spark RDD handles partitioning data across all the nodes in a cluster. It holds them in the memory pool of the cluster as a single unit.
All the functionalities being provided by Apache Spark are built on the top of Spark Core. It manages all essential I/O functionalities. It is used for task dispatching and fault recovery. Spark Core is embedded with a special collection called RDD (Resilient Distributed Dataset). RDD is among the abstractions of Spark. Spark RDD handles partitioning data across all the nodes in a cluster. It holds them in the memory pool of the cluster as a single unit.
attributes
- RDD: distributed data set for paralle computation
- memory sharing between tasks
- broadcast var=sc.broadcast([1,2,3]) print(var.value)
- accumulator
accum=sc.accumulator(0)
sc.parallelize([1,2,3,4]).foreach(lambda x:accum.add(x))
partition structure
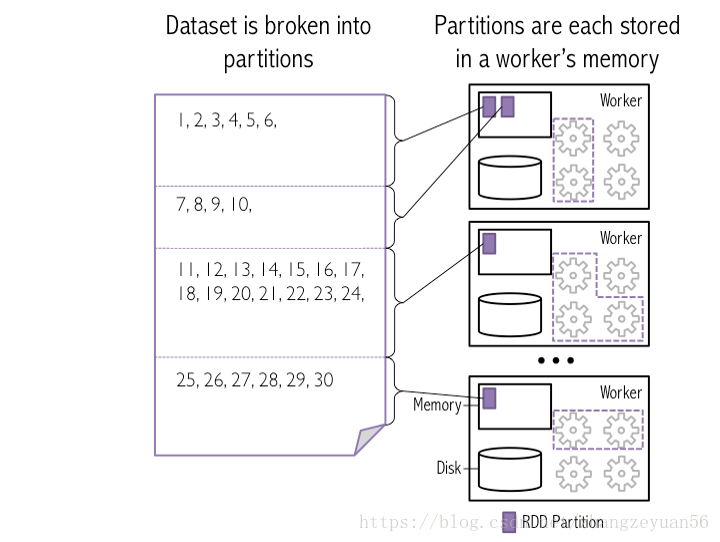 relationship between rdd, partition and task
relationship between rdd, partition and task
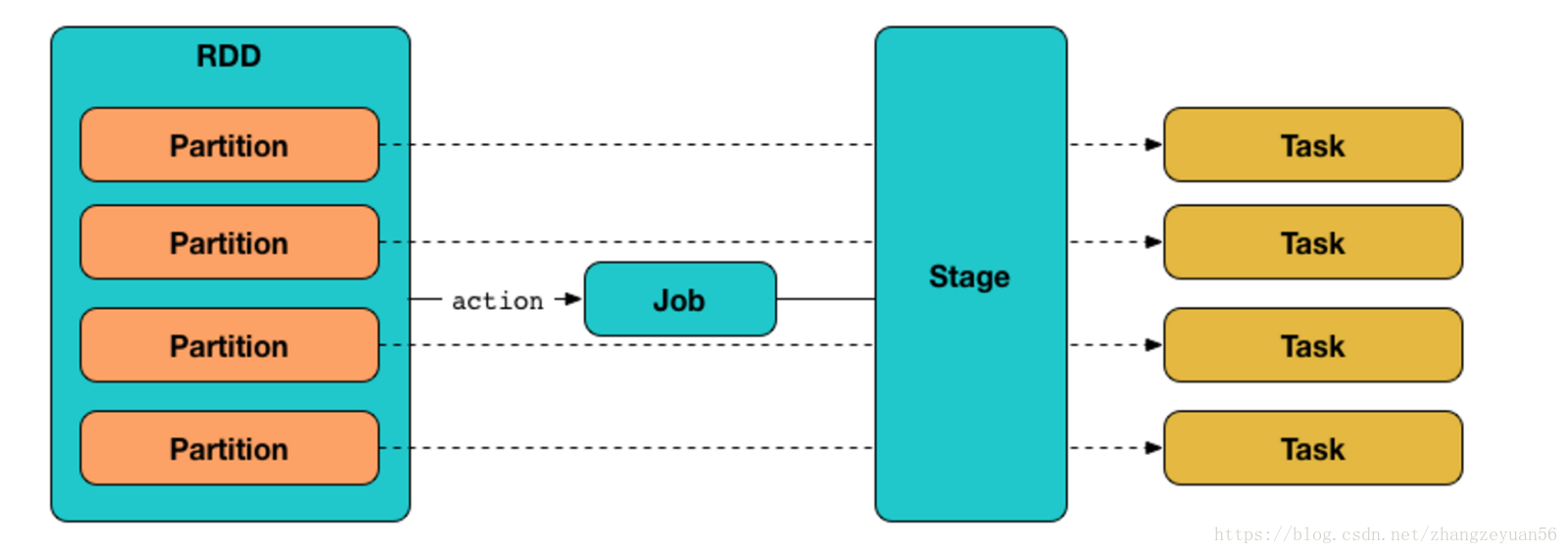
- job is submited when an action is trigered, jobs are done sequentially
- stage: one job contains many stages, stages are seperated by shuffle, which is dependency betwwen parent rdd and children rdd. stages are done sequentially
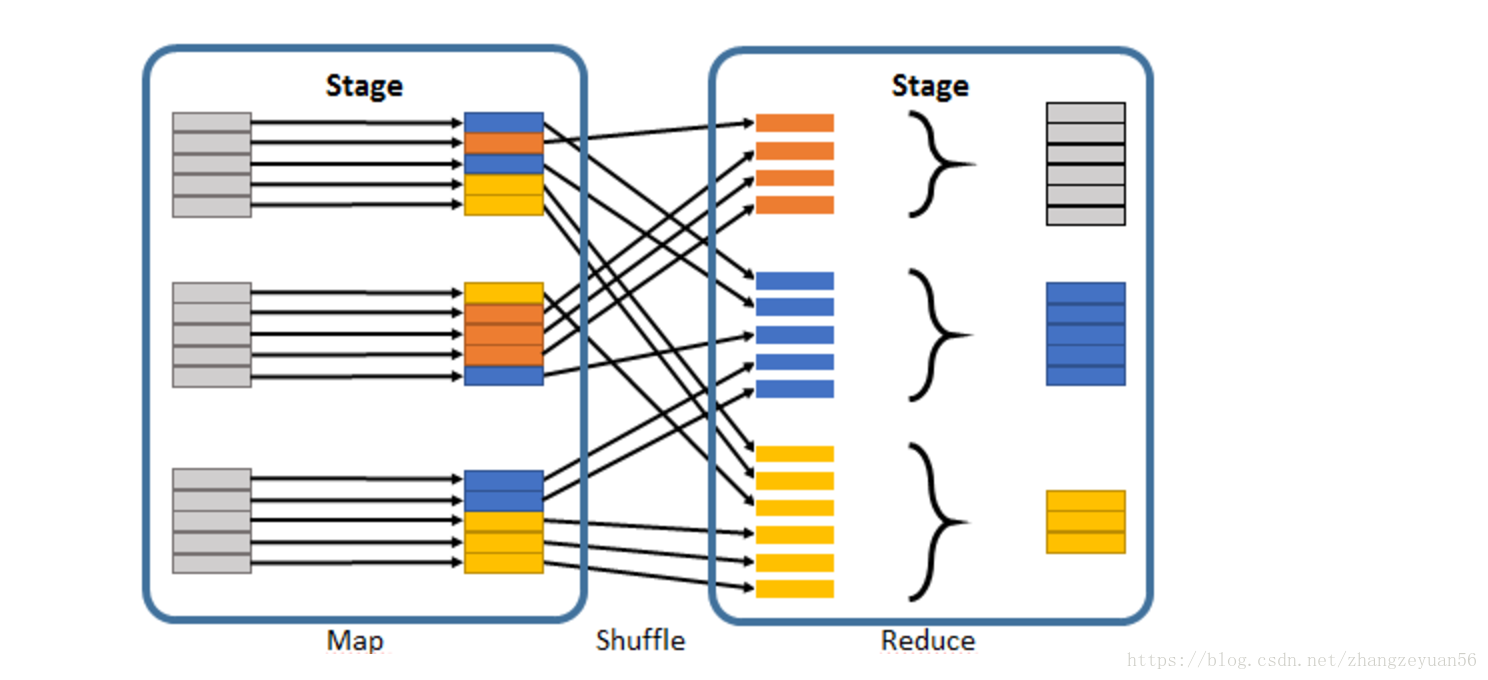
- task: task is the smallest calculation element in spark. the number of tasks is actually the parallelism of stages
partition tuning
The general recommendation for Spark is to have 4x of partitions to the number of cores in cluster available for application. the task should take 100ms+ time to execute
-
Repartition before multiple joins
users = spark.read.load('/path/to/users').repartition('userId') joined1 = users.join(addresses, 'userId') joined1.show() # <-- 1st shuffle for repartition joined2 = users.join(salary, 'userId') joined2.show() # <-- skips shuffle for users since it's already been repartitioned - Repartition after flatMap
- Get rid of disk spills
disk spills result in OutOfMemoryError when one of the reduce tasks in groupByKey was too large
check if disk spilling is occured by searching in log: INFO ExternalSorter: Task 1 force spilling in-memory map to disk it will release 232.1 MB memory solution:- reduce data size
- repartition
- increase shuffle buffer by spark.executor.memory
- reduce number of I/O by spark.shuffle.file.buffer
- data skewness(distribution of data on tasks is not balanced)
- Coalesce after filtering
- Repartition before writing to storage df.write.partitionBy(‘key’).json(‘/path/to/foo.json’)
sparkconf
set configuration for sparks
from pyspark import SparkContext, SparkConf
conf=SparkConf()
conf.setMaster('local').setAppName('My App') #local is only for spark computation on local machine. if on cluster then leave it out, this parameter will be passed in command line with '--master' option
sparkcontext
it is an interface to call APIs in spark
sc=SparkContext(conf=conf)
RDD
RDD is an abstract data type in spark, is similar to array. It is stored partitionedly
-
using parallelied aggregation
rdd=sc.parallelize(range(9),1) print(rdd.collect())
-
using local file or hdfs
lines=sc.textFile(path) words=lines.flatMap(lambda line:line.split(" ")) keyvalue=words.map(lambda word:(word,1)) print(keyvalue.countByKey())
DataFrame
read dataframes
#read csv as DataFrame
df = spark.read.load("filename.csv",format="csv", sep=",", inferSchema="true", header="true")
#or
df = spark.read.csv("filenames.csv", header="true", inferSchema="true") ### preprocessing
#remove unnecessary column
df = df.drop("instant")
#print schema of dataset
df.printSchema()
#show column names
df.columns
#show counts of rows and columns
df.count()
len(df.columns)
#show describe information
df.describe('Glucose').show()
using SQL
Spark SQL can be used to enquire structured data stored in Hadoop and Spark cluster, it is used to evaluates the values of a specific column and changes the value to something more meaningful.
#evaluation and categorize data
from pyspark.sql.functions import col,when
df_when = df.withColumn("BloodPressure", when(col("BloodPressure") >125,"High")
.when(col("BloodPressure") <80,"Low")
.otherwise("Normal"))
df_when.show() ### run sql query run any SQL queries within pySpark and the result will be in form of a dataframe
df.createOrReplaceTempView("diabetesstudy")
sqldf = spark.sql("SELECT * FROM diabetesstudy")
sqldf.show(10)
#or register df as sql table
df.registerTempTable('cases_table')
df_new = sqlContext.sql('select * from cases_table where confirmed>100')
create coloumn
import pyspark.sql.functions as F
rdd_new = rdd.withColumn("NewConfirmed", 100 + F.col("confirmed"))
convert dataframe between rdd
#convert df to rdd
df.rdd
#convert rdd to df
sqlContext.createDataFrame(rdd)
UDF
define a normal function in python and use it in pyspark
import pyspark.sql.functions as F
from pyspark.sql.types import *
def casesHighLow(confirmed):
if confirmed < 50:
return 'low'
else:
return 'high'
#convert to a UDF Function by passing in the function and return type of function
casesHighLowUDF = F.udf(casesHighLow, StringType())CasesWithHighLow = cases.withColumn("HighLow", casesHighLowUDF("confirmed"))
using pandas in spark
a decorator F.pandas_udf and an output shema need to be defined
spark windows functions
from pyspark.sql.window import Window
windowSpec = Window().partitionBy(['province']).orderBy(F.desc('confirmed'))
cases.withColumn("rank",F.rank().over(windowSpec)).show()
Pivot Dataframes
pivotedTimeprovince = timeprovince.groupBy('date').pivot('province').agg(F.sum('confirmed').alias('confirmed') , F.sum('released').alias('released'))
pivotedTimeprovince.limit(10).toPandas()
transformation
-
map processing input with specific operation and return result object
rdd=sc.parallelize(range(9),1)
print(rdd.map(lambda x:x).collect())
print(rdd.map(lambda x:x+1).collect()) -
mapPartitions(func, preservesPartitioning=False)
applying func on every slice rdd -
flatMap
map + flatten(concatenate all objects to one objects)rdd=sc.parallelize([1,2,3])
print(rdd.map(lambda x: range(x)).collect())
print(rdd.flatMap(lambda x: range(x)).collect()) -
flatMapValues(f)
flat the value map without changing the keydef func(x): return x
sc.parallelize([(“a”,[“x”,”y”,”z”]), (“b”,[“p”,”r”])])
flatMapValues(func).collect() -
mapValues processing value with an operation
rdd=sc.parallelize([(“python”, 1), (“c++”, 2)])
print(rdd.map(lambda x:x).collect())
print(rdd.mapValues(lambda x:x*2).collect() ) -
filter filter with condition
rdd.filter(lambda x:x%2==0).collect() #in spark dataframe df.filter((rdd.confirmed>10) & (rdd.province==’Daegu’))
-
distinct remove duplicates
#drop duplicates considering all columns rdd.distinct().collect() #drop duplicates considering only a subset of the columns #the returned DataFrame will contain only the subset of the columns that was used to eliminate the duplicates rdd.select([‘id’, ‘name’]).distinct().collect()
-
dropDuplicates drop duplicates over all or a subset of columns, and keep all the columns in the returned dataframe
df.dropDuplicates([‘id’, ‘name’]).show()
-
randomSplit slice data with portion for test and train purposes
rddsplitted=rdd.randomsplit([0.2, 0.6])
print(rddsplitted[0].collect()) -
groupBy grouping by condition
rddgrouped=rdd.groupBy(lambda x: “less” if(x<10) else “more”)
print(rddgrouped.mapValues(list).collect()) # in dataframe
df.groupBy([“province”,”city”]).agg(F.sum(“confirmed”)).show() df.groupBy([“province”,”city”]).agg(F.sum(“confirmed”).alias(“TotalConfirmed”)).show() -
groupbykey group by key
-
reduceByKey processing element with same key with one operation
rdd.reduceByKey(add).collect()
-
sortBy(keyfunc, ascending=True, numPartitions=None) sorting rdd with defined element( key or value)
rdd.sortBy(lambda x: x[0]).collect() # sort by key
rdd.sortBy(lambda x: x[1]).collect() # sort by value -
sortByKey
-
aggregate(zeroValue, seqOp, combOp) process zeroValue and element in rdd in first slice with seqOp, then operate results data from each slice with combOp
seqOp=(lambda x,y: (x[0]+y, x[1]+1))
combOp=(lambda x,y: (x[0]+y[0], x[1]+y[1]))
sc.parallelize([1,2,3,4]).aggregate((0,0), seqOp, comOp) #in dataframe #joining dataframe with different size from pyspark.sql.functions import broadcast df1.join(broadcast(df2), coloumn_name_list,how=’left’) -
aggregateByKey(zeroValue, seqOp, combOp, numPartitions=None, partitionFunc=None)
aggregate value when key is the same and additionally zeroValue is added to the valueseqOp=(lambda x,y: x+y)
combOp=(lambda x,y: x+y)
sc.parallelize([(1,2),(1,3),(1,4)]).aggregateByKey(3,seqOp, combOp).collect() -
join(rdd, numPartitions=None)
combining value in list with same keyrdd1=sc.parallelize([(“a”, 1), (“b”,1)])
rdd2=sc.parallelize([(“a”, 3), (“b”,4)])
rdd1.join(rdd2).collect()
# in dataframe rdd1.join(rdd2, coloumn_name_list,how=’left’) -
like similar to the like filter in SQL. ‘%’ can be used as a wildcard to filter the result
df.select(“Pregnancies”,”Glucose”,df.BMI.like(‘33%’)).show(10)
action
it get elements in rdd, return to drive and triger the spark job and transformation
-
reduce
reduce dimension with an operationrdd.reduce(add)
-
collect
get the list of all elements in rdd to local client -
count count number of elements in list
-
take(n)
get first n elements from rdd -
first()
get first element from rdd -
top(n)
get n max elements from rdd -
takeOrdered(n, [, key=None])
get first n elements from rdd after sorting - min, max, mean, stdev
-
fold
initialize zeroValue in each slice and let zeroValue participat in op calculation, at the end the result in every slice will be combined into one slice(zeroValue also participate in calculation)addOp=(lambda x,y:x+y)
rdd1=sc.parallelize(range(6), 1)
rdd2=sc.parallelize(range(6),2) print(rdd1.fold(1,addOp))
print(rdd2.fold(1,addOp)) - countByKey()
count elements with same key - countByValue()
count elements with same value -
takeSample(bool,n)
bool:true->sampling with replacement; false->sampling without replacement
n: number of sample to be takenrdd.takeSample(true, 4)
-
foreach or items()
loop through all element in rdd -
glom()
combining elements in different slice into one rdd list - saveAsTextFile
save elements in rdd to file -
textFile
load text to memorysc.textFile(path)
-
write.csv(path, mode=”overwrite”)
-
toPandas()
convert spark dataframe to pandas dataframe -
withColumnRenamed(old_name, new_name) or toDF(all_new_coloumn_name_lists)
-
select(coloumn_name)
select specified coloumn with name - withColumn(coloumn_name, F.col(coloumn_name).cast(IntegerType()))
cast data in specified coloumn to other type
F is imported: from pyspark.sql import functions as F to cast data type for all column: df = df.select([F.col(c).cast(“double”).alias(c) for c in df.columns])
shuffle
the process to gather datas on different nodes to one node with specific rule is called shuffle
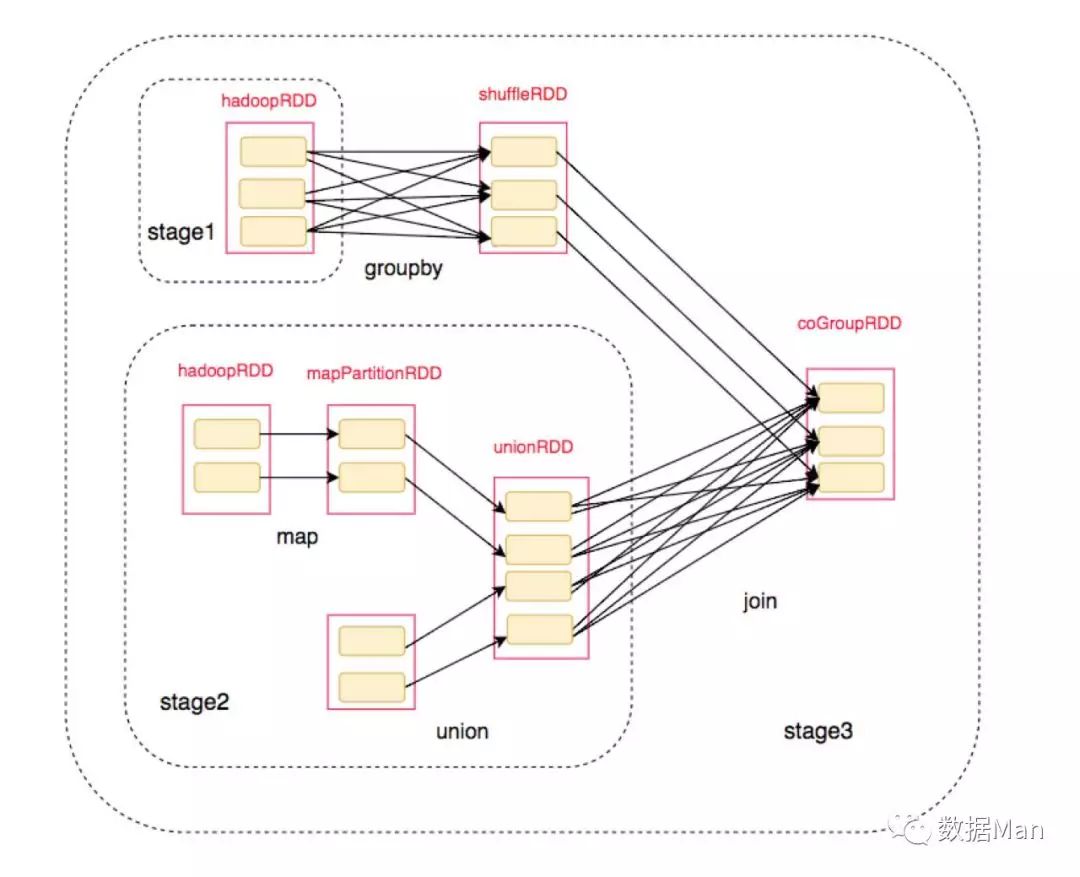
- shuffle read is also called Map Task in mapReduce Shuffle
- shuffle write is also called Reduce Task in mapReduce Shuffle
persist
-
persist() or cache() store transformation result in memory for second usage
variable.persist() # keep this variable in memory #or variable.cache()
-
unpersist() variable.unpersist()
Tips
Partitioning and resource management
- activating dynamic allocation of executors via the spark.dynamicAllocation.maxExecutors and spark.dynamicAllocation.enabled parameters can considerably decrease idleness of Spark’s computational resources
- Splitting a large processing job into multiple smaller jobs. For each of these smaller jobs, we can also set the parameters spark.default.parallelism and spark.sql.shuffle.partitions appropriately to prevent the need for constant re-partitioning.
Immutability, lazy evaluation and execution plan optimization
Transformations produce a new Spark dataset as output (Spark has the immutability property so it can never modify existing datasets, only create new ones); Actions take Spark datasets as inputs but result in something else than a Spark dataset, such as writing into storage, creating a local (non-Spark) variable or displaying something in the user’s UI.
Transformations are being done at partition level, not dataset level
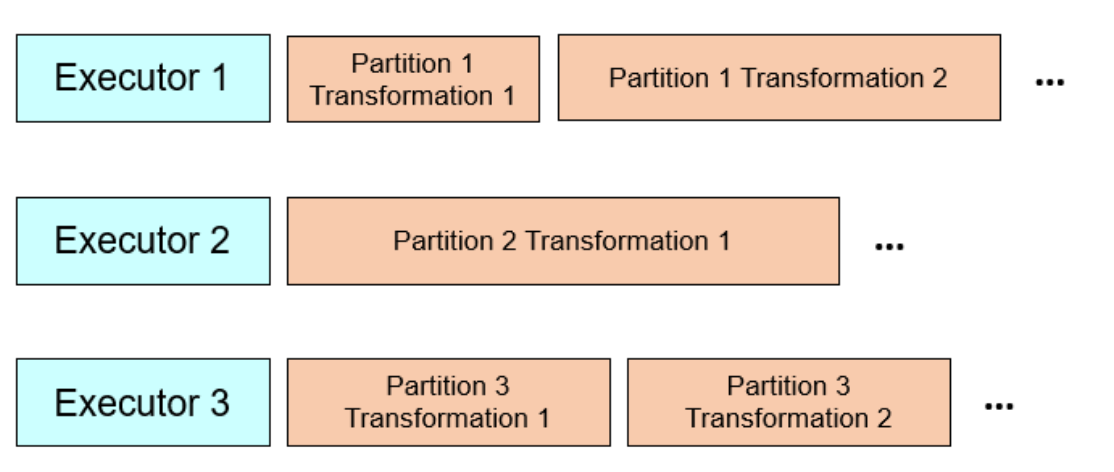 Spark optimizes execution plans, and the larger the execution plans, the better for optimization
Spark optimizes execution plans, and the larger the execution plans, the better for optimization
pyspark programming
run spark python script
spark-submit script.py
pyspark jupyter-notebook
#add this function to .bashrcs
function pysparknb ()
{
#Spark path
SPARK_PATH=spark_path_folder
export PYSPARK_DRIVER_PYTHON="jupyter"
export PYSPARK_DRIVER_PYTHON_OPTS="notebook"
# For pyarrow 0.15 users, you have to add the line below while using pandas_udf
export ARROW_PRE_0_15_IPC_FORMAT=1# Change the local[10] to local[numCores in your machine]
$SPARK_PATH/bin/pyspark --master local[10]
}
pySpark query plan
query = (
questionsDF
.filter(col('year') == 2019)
.groupBy('user_id')
.agg(
count('*').alias('cnt')
)
.join(usersDF, 'user_id')
)
query.explain(mode='formatted')
pySpark ML
MLlib is Spark’s machine learning (ML) library
ML Algorithms
from pyspark.ml.feature import VectorAssembler
#Vector Assemble is an important method. It is used for assembling all the features in one vector
assembler = VectorAssembler(inputCols=['Glucose', 'BloodPressure', 'BMI', 'Age'], outputCol='DiabFeature')
df_ml=df.select("Glucose","BloodPressure","BMI","Age","Outcome")
df_transform = assembler.transform(df_ml)
df_transform.show(10)
#Split the data in train and test
(dftrain, dftest) = df_transform.randomSplit([0.8,0.2], seed =2020)
print("Train data count: ", dftrain.count())
print("Test data count: ", dftest.count())
from pyspark.ml.classification import GBTClassifier
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
gb = GBTClassifier(labelCol = 'Outcome', featuresCol = 'DiabFeature')
gbModel = gb.fit(dftrain)
gb_predictions = gbModel.transform(dftest)
##Evaluat the performance of model
multi_evaluator = MulticlassClassificationEvaluator(labelCol = 'Outcome', metricName = 'accuracy')
print('Gradient-boosted Trees Accuracy:', multi_evaluator.evaluate(gb_predictions))
Pipelines
from pyspark.ml.feature import VectorAssembler, VectorIndexer
featuresCols = df.columns
featuresCols.remove('cnt')
# Concatenates all feature columns into a single feature vector in a new column "rawFeatures"
vectorAssembler = VectorAssembler(inputCols=featuresCols, outputCol="rawFeatures")
# Identifies categorical features and indexes them
vectorIndexer = VectorIndexer(inputCol="rawFeatures", outputCol="features", maxCategories=4)
from pyspark.ml.regression import GBTRegressor
# Takes the "features" column and learns to predict "cnt"
gbt = GBTRegressor(labelCol="cnt")
from pyspark.ml.tuning import CrossValidator, ParamGridBuilder
from pyspark.ml.evaluation import RegressionEvaluator# Define a grid of hyperparameters to test:
# - maxDepth: max depth of each decision tree in the GBT ensemble
# - maxIter: iterations, i.e., number of trees in each GBT ensemble
# In this example notebook, we keep these values small. In practice, to get the highest accuracy, you would likely want to try deeper trees (10 or higher) and more trees in the ensemble (>100)paramGrid = ParamGridBuilder()\
.addGrid(gbt.maxDepth, [2, 5])\
.addGrid(gbt.maxIter, [10, 100])\
.build()
# We define an evaluation metric. This tells CrossValidator how well we are doing by comparing the true labels with predictions.
evaluator = RegressionEvaluator(metricName="rmse", labelCol=gbt.getLabelCol(), predictionCol=gbt.getPredictionCol())
# Declare the CrossValidator, which runs model tuning for us.
cv = CrossValidator(estimator=gbt, evaluator=evaluator, estimatorParamMaps=paramGrid)
# tie feature and training model in the pipeline
from pyspark.ml import Pipeline
pipeline = Pipeline(stages=[vectorAssembler, vectorIndexer, cv])
# Train & Test
pipelineModel = pipeline.fit(train)
predictions = pipelineModel.transform(test)
rmse = evaluator.evaluate(predictions)
Spark Delight
the Spark Delight aims to solve the weakness of spark UI and Spark History Server:
- The Spark UI lacks essential node metrics (CPU, Memory and I/O usage)
- The Spark History Server (rendering the Spark UI after an application is finished) is hard to setup.
Summary statistics
shows statistical performance of spark app
Recommendations
pinpoints stability and performance issues at a high-level to help developers address them
Executors CPU Usage
let you see quickly if your app is I/O bound or CPU bound, and make smarter infrastructure changes accordingly
Executors Peak Memory Usage
shows the memory usage breakdown for each executor when the total memory consumption was at its peak
Stage and Executor Pages
show information at a finer granularity on a Spark stage page or an executor page